Visualization
Jeff Goldsmith, Fabian Scheipl
2024-02-23
Source:vignettes/x04_Visualization.Rmd
x04_Visualization.RmdThe tidyfun package is designed to
facilitate functional data analysis in R,
with particular emphasis on compatibility with the
tidyverse. In this vignette, we illustrate data
visualization using tidyfun.
We’ll draw on tidyfun::chf_df and
tidyfun::dti_df, as well as the
fda::CanadianWeather data.
Plotting with ggplot
ggplot is a powerful framework for visualization. In
this section, we’ll assume some basic familiarity with the package; if
you’re new to ggplot, this primer may be
helpful.
tidyfun includes
pasta-themed geoms and plots for
functional data:
-
geom_spaghettifor lines -
geom_meatballsfor (lines &) points -
gglasagnafor heatmaps, with anorder-argument for arranging lasagna layers / heat map rows. -
geom_capellinifor little sparklines / glyphs on maps etc. -
geom_errorband– a functional data version ofgeom_ribbon
geom_spaghetti and geom_meatballs
One of the most fundamental plots for functional data is the
spaghetti plot, which is implemented in tidyfun +
ggplot through geom_spaghetti:
chf_df |>
filter(id == 1) |>
ggplot(aes(y = activity)) +
geom_spaghetti()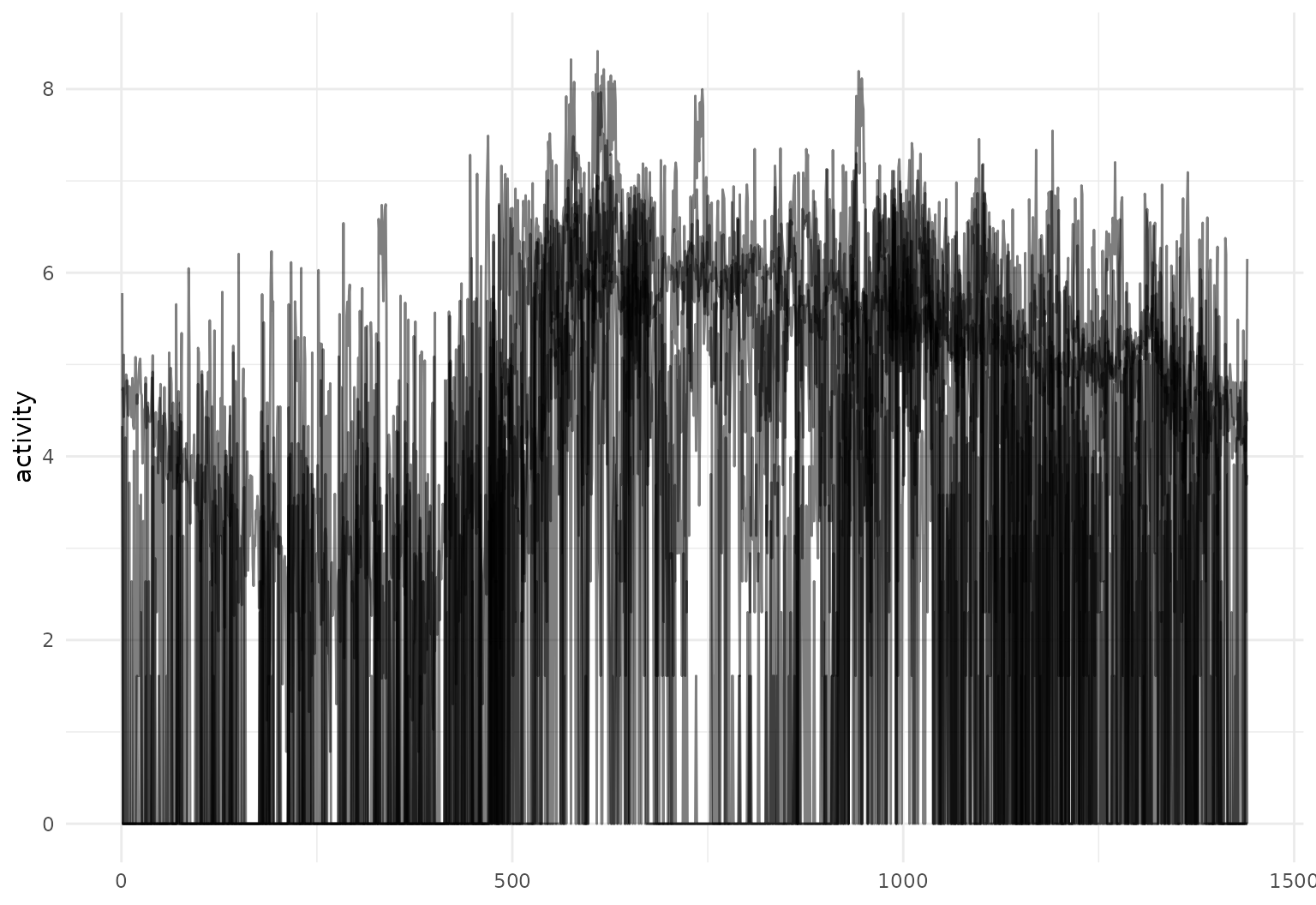
A variant on the spaghetti plot is the meatballs plot, which shows both the “noodles” (i.e. functional observations visualized as curves) and the “meatballs” (i.e. observed data values visualized as points).
chf_df |>
filter(id == 1, day == "Mon") |>
ggplot(aes(y = activity)) +
geom_meatballs()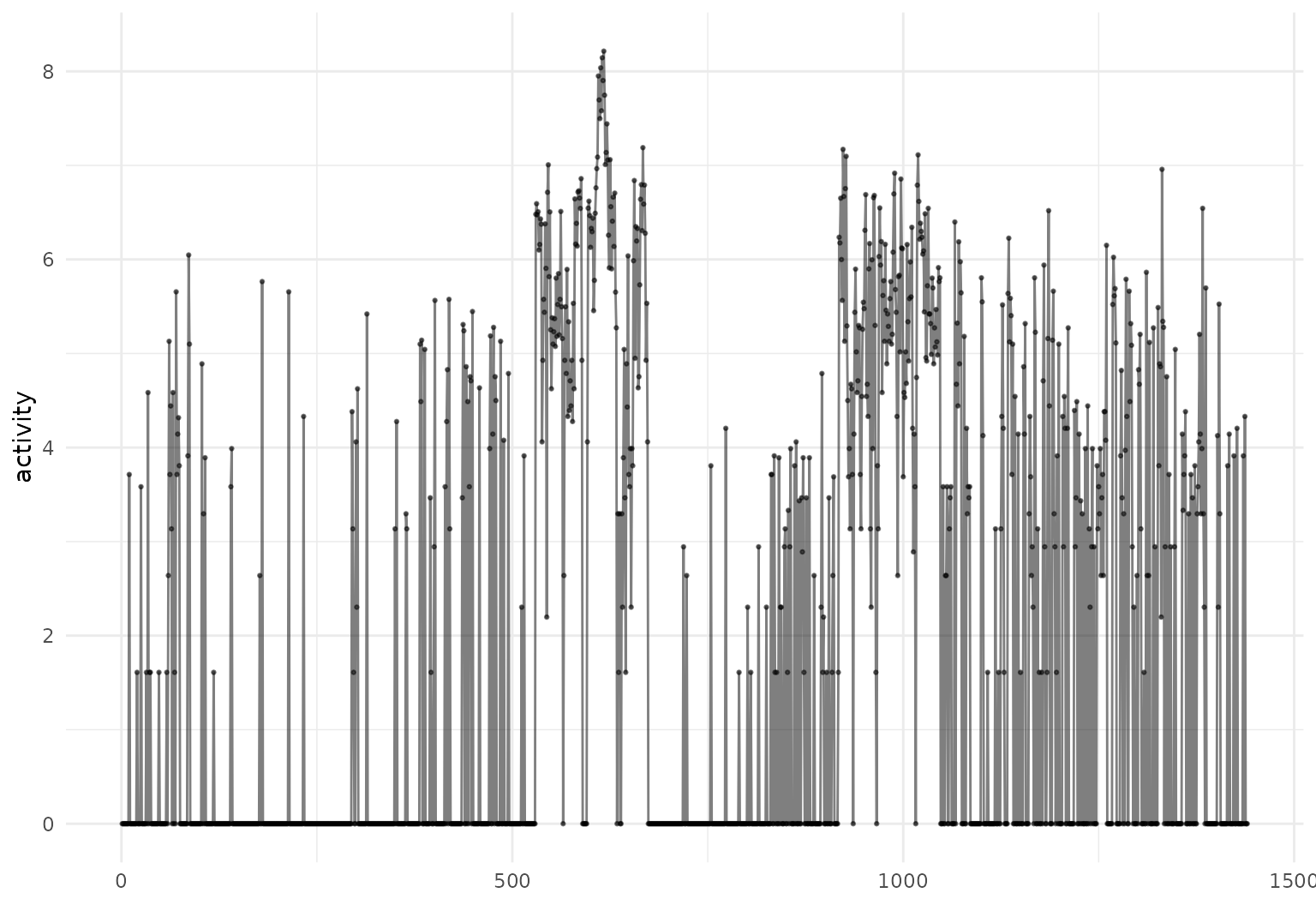
Using with other ggplot features
The new geoms in tidyfun “play nicely” with standard
ggplot aesthetics and options.
You can, for example, define the color aesthetic for plots of
tf variables using other observations:
chf_df |>
filter(id %in% 1:5) |>
ggplot(aes(y = activity, color = gender)) +
geom_spaghetti(alpha = 0.2)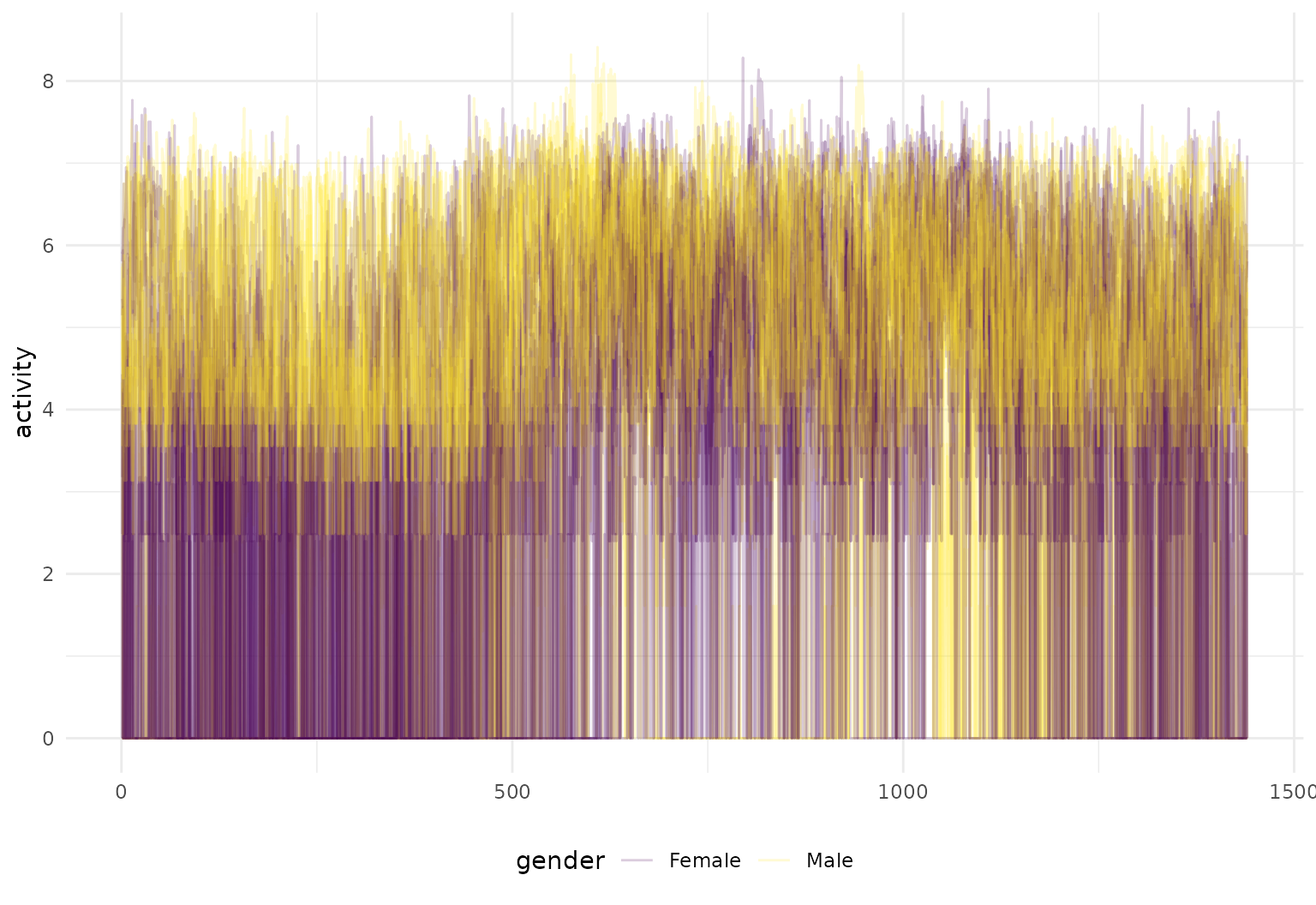
You can also use facetting:
chf_df |>
filter(day %in% c("Mon", "Sun")) |>
ggplot(aes(y = activity, color = gender)) +
geom_spaghetti(alpha = 0.1) +
facet_grid(~day)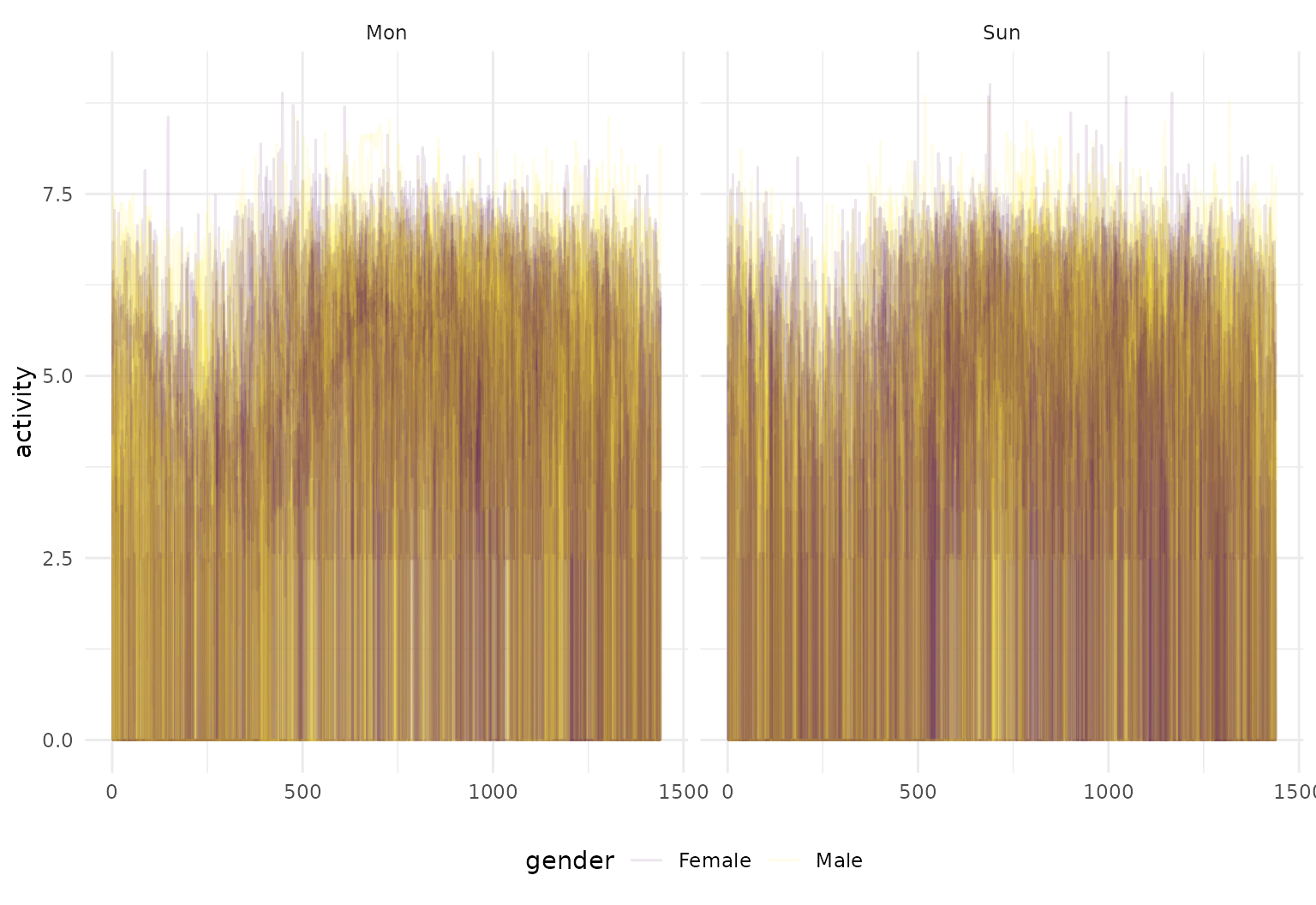
Another example, using the DTI data, is below.
dti_df |>
ggplot() +
geom_spaghetti(aes(y = cca, col = case, alpha = 0.2 + 0.4 * (case == "control"))) +
facet_wrap(~sex) +
scale_alpha(guide = "none", range = c(0.2, 0.4))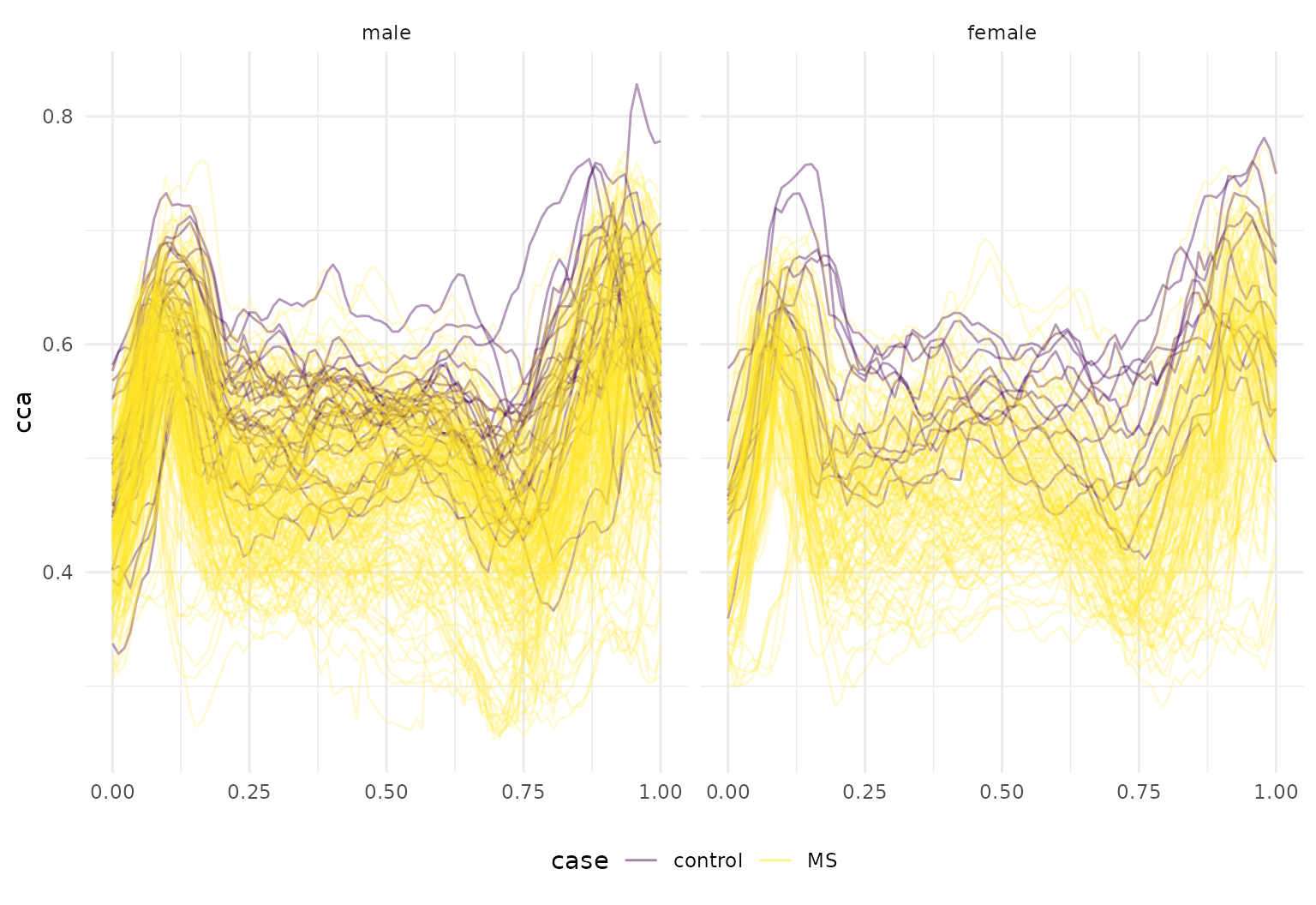
Together with tidyfun’s tools for
functional data wrangling and summary statistics, the integration with
ggplot2 can produce useful exploratory analyses, like the
plot below showing group-wise smoothed and unsmoothed mean activity
profiles:
chf_df |>
group_by(gender, day) |>
summarize(mean_act = mean(activity)) |>
mutate(smooth_mean = tfb(mean_act, verbose = FALSE)) |>
filter(day %in% c("Mon", "Sun")) |>
ggplot(aes(y = smooth_mean, color = gender)) +
geom_spaghetti(linewidth = 1.25, alpha = 1) +
geom_meatballs(aes(y = mean_act), alpha = 0.1) +
facet_grid(~day)
## `summarise()` has grouped output by 'gender'. You can override using the
## `.groups` argument.
## Percentage of input data variability preserved in basis representation (per
## functional observation, approximate): Min. 1st Qu. Median Mean 3rd Qu. Max.
## 100 100 100 100 100 100
## Percentage of input data variability preserved in basis representation (per
## functional observation, approximate): Min. 1st Qu. Median Mean 3rd Qu. Max.
## 100 100 100 100 100 100
## Percentage of input data variability preserved in basis representation (per
## functional observation, approximate): Min. 1st Qu. Median Mean 3rd Qu. Max.
## 100 100 100 100 100 100
## Percentage of input data variability preserved in basis representation (per
## functional observation, approximate): Min. 1st Qu. Median Mean 3rd Qu. Max.
## 100 100 100 100 100 100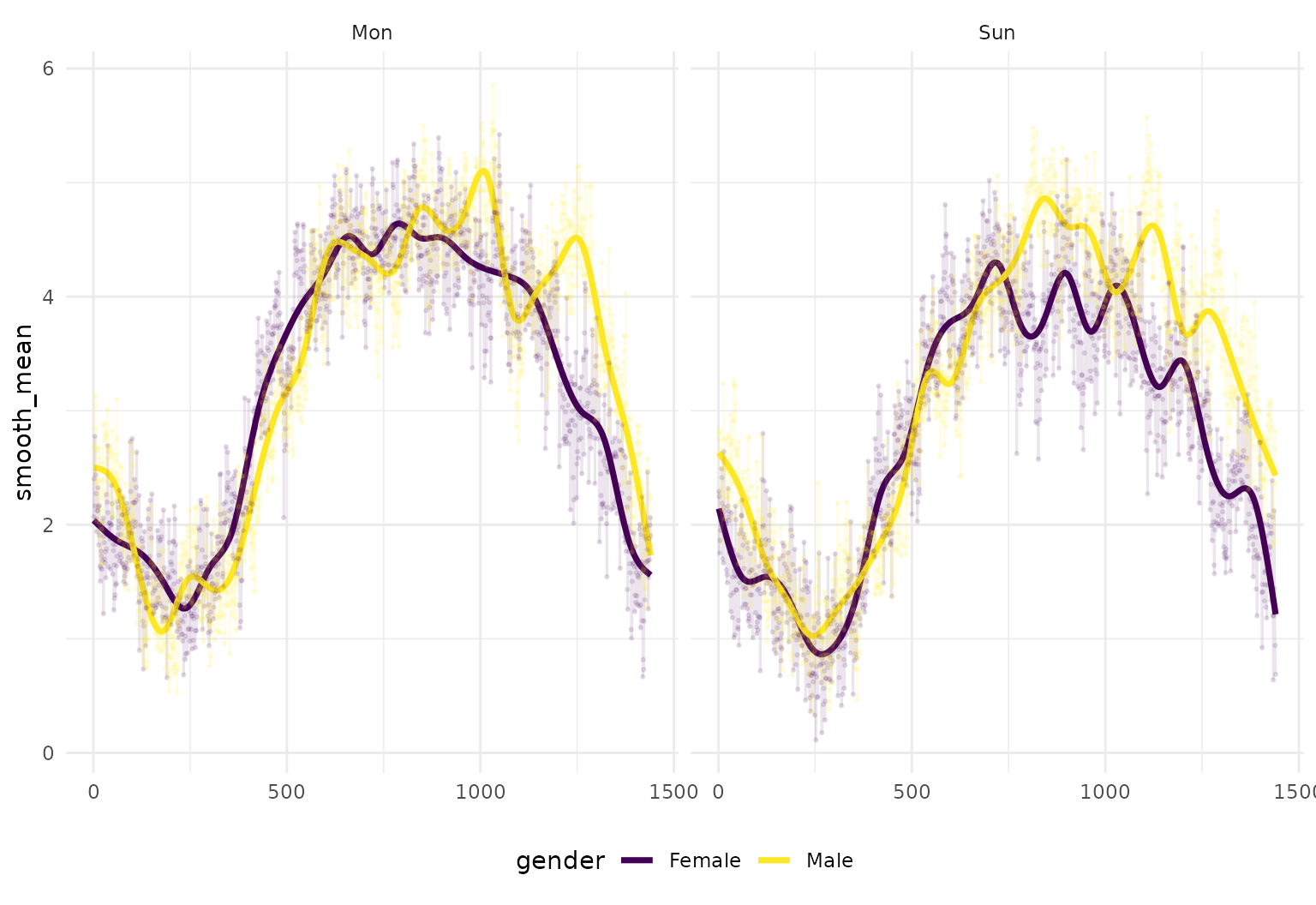
… or the plot below showing group-wise mean functions +/- twice their pointwise standard errors:
chf_df |>
group_by(gender, day) |>
summarize(
mean_act = mean(activity),
sd_act = sd(activity)
) |>
group_by(gender, day) |>
mutate(
upper_act = mean_act + 2 * sd_act,
lower_act = mean_act - 2 * sd_act
) |>
filter(day %in% c("Mon", "Sun")) |>
ggplot(aes(y = mean_act, color = gender, fill = gender)) +
geom_spaghetti(alpha = 1) +
geom_errorband(aes(ymax = upper_act, ymin = lower_act), alpha = 0.3) +
facet_grid(day ~ gender)
## `summarise()` has grouped output by 'gender'. You can override using the
## `.groups` argument.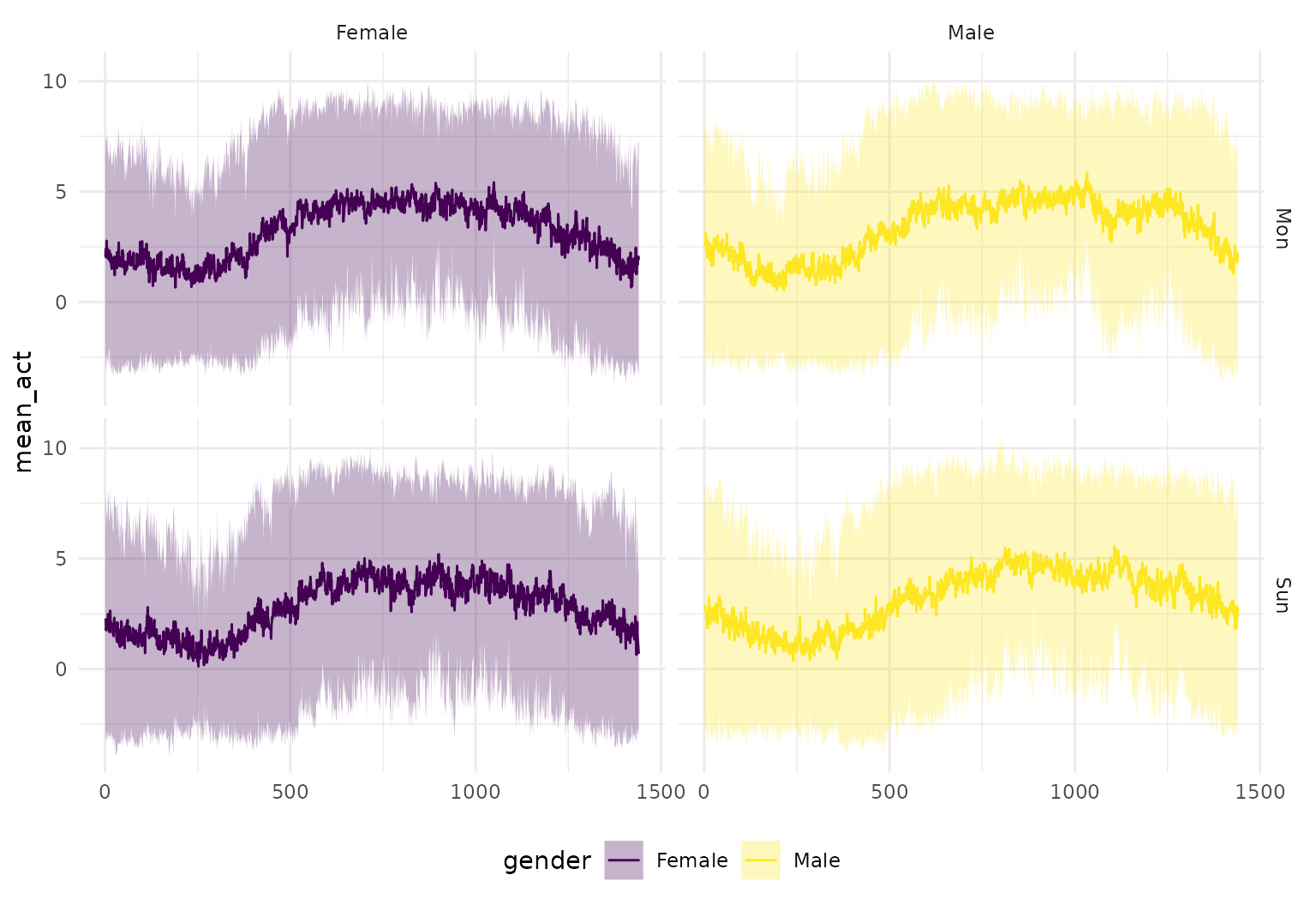
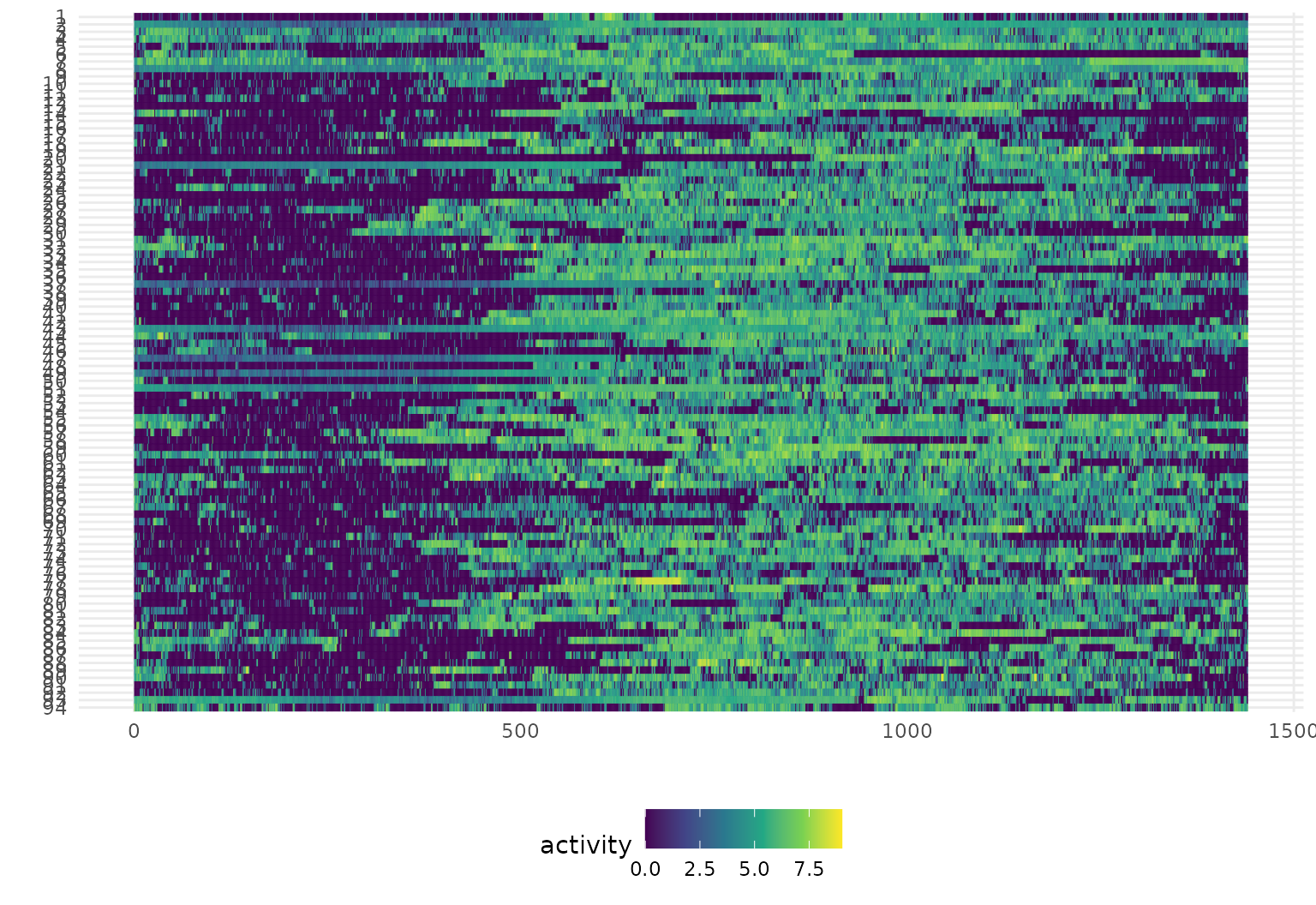
gglasagna
Lasagna plots are “a saucy alternative to spaghetti plots”. They are a variant on a heatmaps which show functional observations in rows and use color to illustrate values taken at different arguments.
In tidyfun, lasagna plots are implemented through
gglasagna. A first example, using the CHF data, is
below.
A somewhat more involved example, demonstrating the
order argument and taking advantage of facets, is next.
dti_df |>
gglasagna(
tf = cca,
order = tf_integrate(cca, definite = TRUE),
arg = seq(0, 1, length.out = 101)
) +
theme(axis.text.y = element_text(size = 6)) +
facet_wrap(~case, ncol = 2, scales = "free")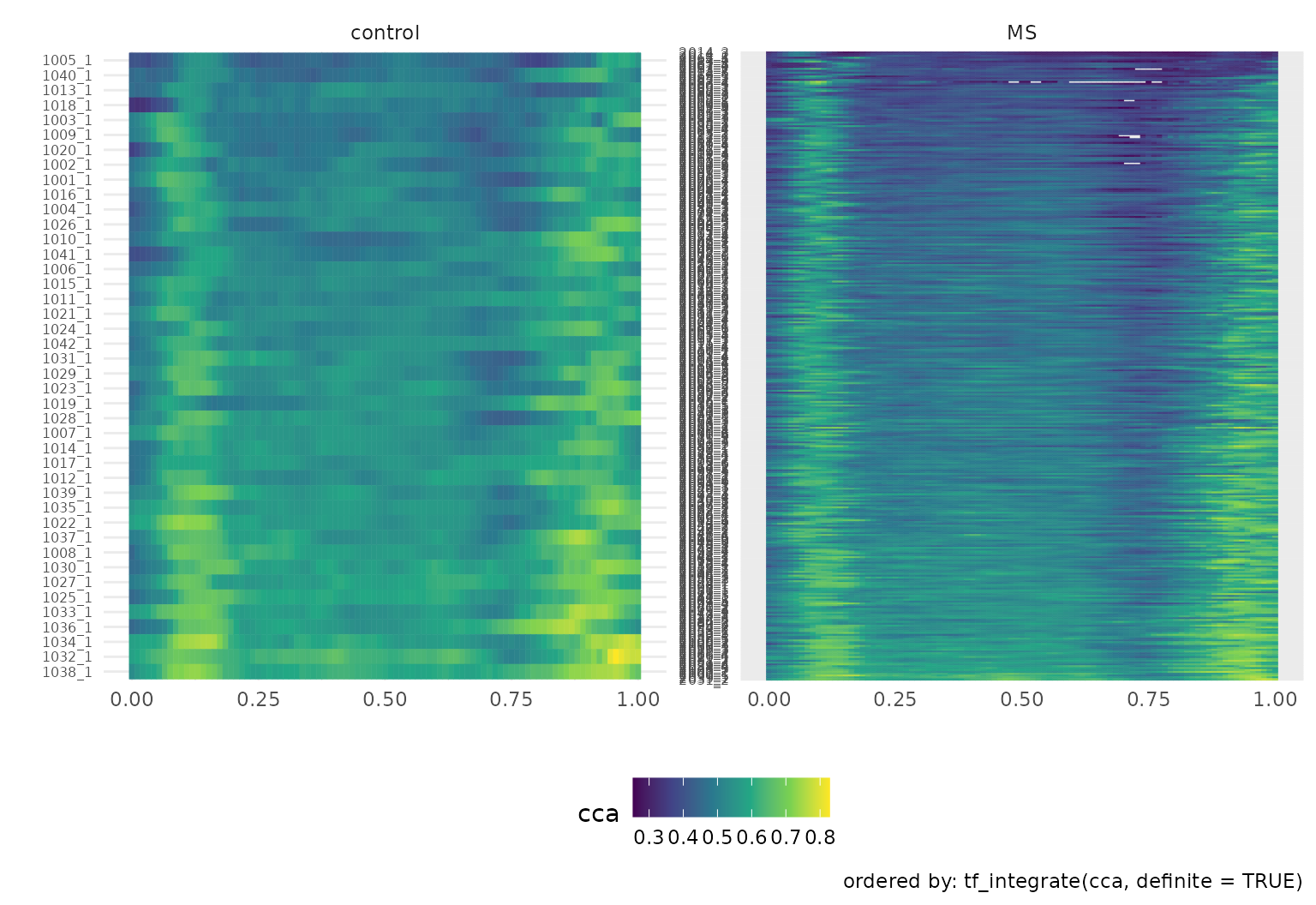
geom_capellini
To illustrate geom_capellini, we’ll start with some data
prep for the iconic Canadian Weather data:
canada <- data.frame(
place = fda::CanadianWeather$place,
region = fda::CanadianWeather$region,
lat = fda::CanadianWeather$coordinates[, 1],
lon = -fda::CanadianWeather$coordinates[, 2]
)
canada$temp <- tfd(t(fda::CanadianWeather$dailyAv[, , 1]), arg = 1:365)
canada$precipl10 <- tfd(t(fda::CanadianWeather$dailyAv[, , 3]), arg = 1:365) |>
tf_smooth()
## using f = 0.15 as smoother span for lowess
canada_map <-
data.frame(maps::map("world", "Canada", plot = FALSE)[c("x", "y")])Now we can plot a map of Canada with annual temperature averages in red, precipitation in blue:
ggplot(canada, aes(x = lon, y = lat)) +
geom_capellini(aes(tf = precipl10),
width = 4, height = 5, colour = "blue",
line.linetype = 1
) +
geom_capellini(aes(tf = temp),
width = 4, height = 5, colour = "red",
line.linetype = 1
) +
geom_path(data = canada_map, aes(x = x, y = y), alpha = 0.1) +
coord_quickmap()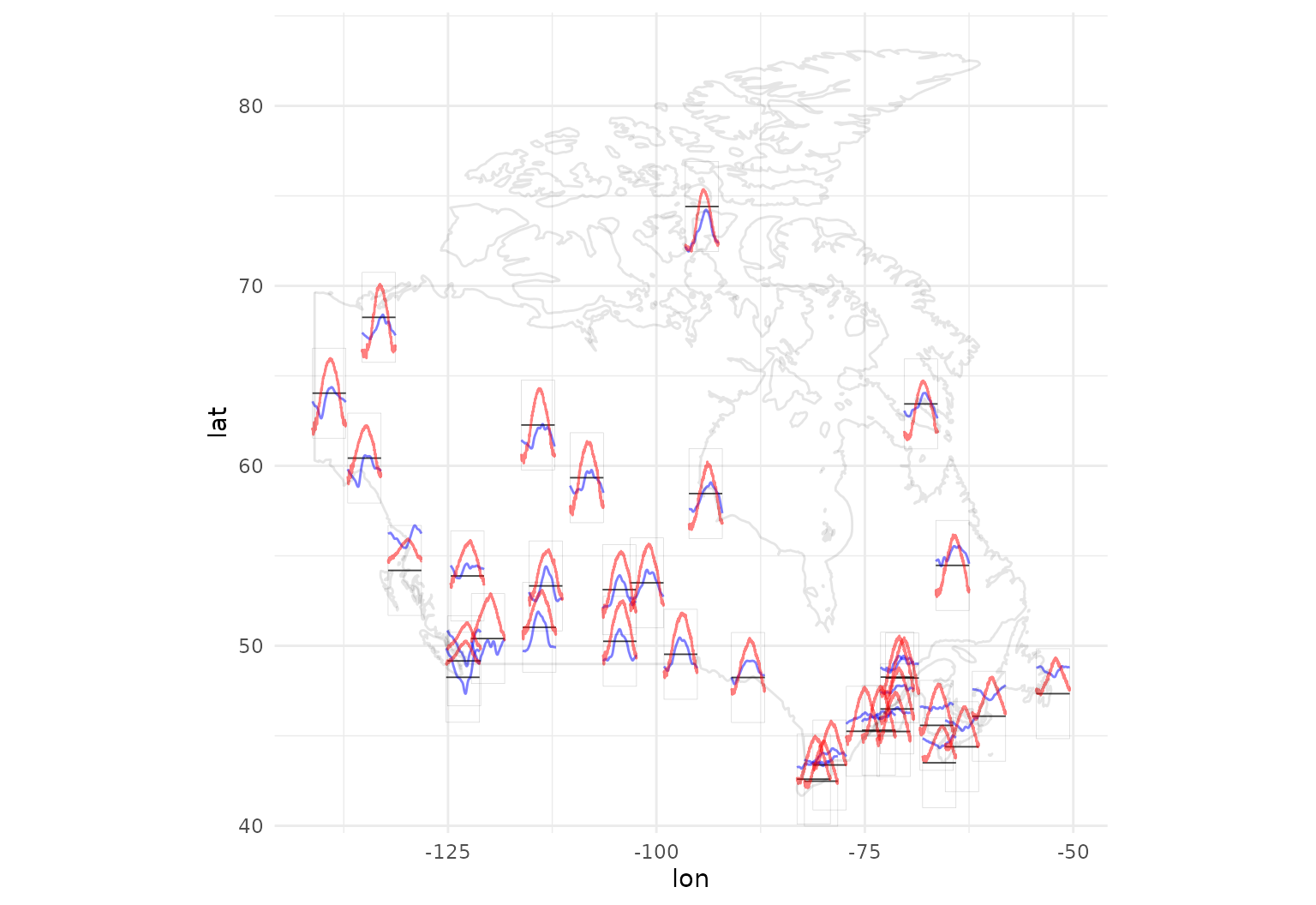
Plotting with base R
tidyfun includes several extensions of
base R graphics, which operate on tf vectors. For example,
one can use plot to create either spaghetti or lasagna
plots, and lines to add lines to an existing plot:
cca <- dti_df$cca |>
tfd(arg = seq(0, 1, length.out = 93), interpolate = TRUE)
layout(t(1:2))
plot(cca, type = "spaghetti")
lines(c(median(cca), mean = mean(cca)), col = c(2, 4))
plot(cca, type = "lasagna", col = viridis(50))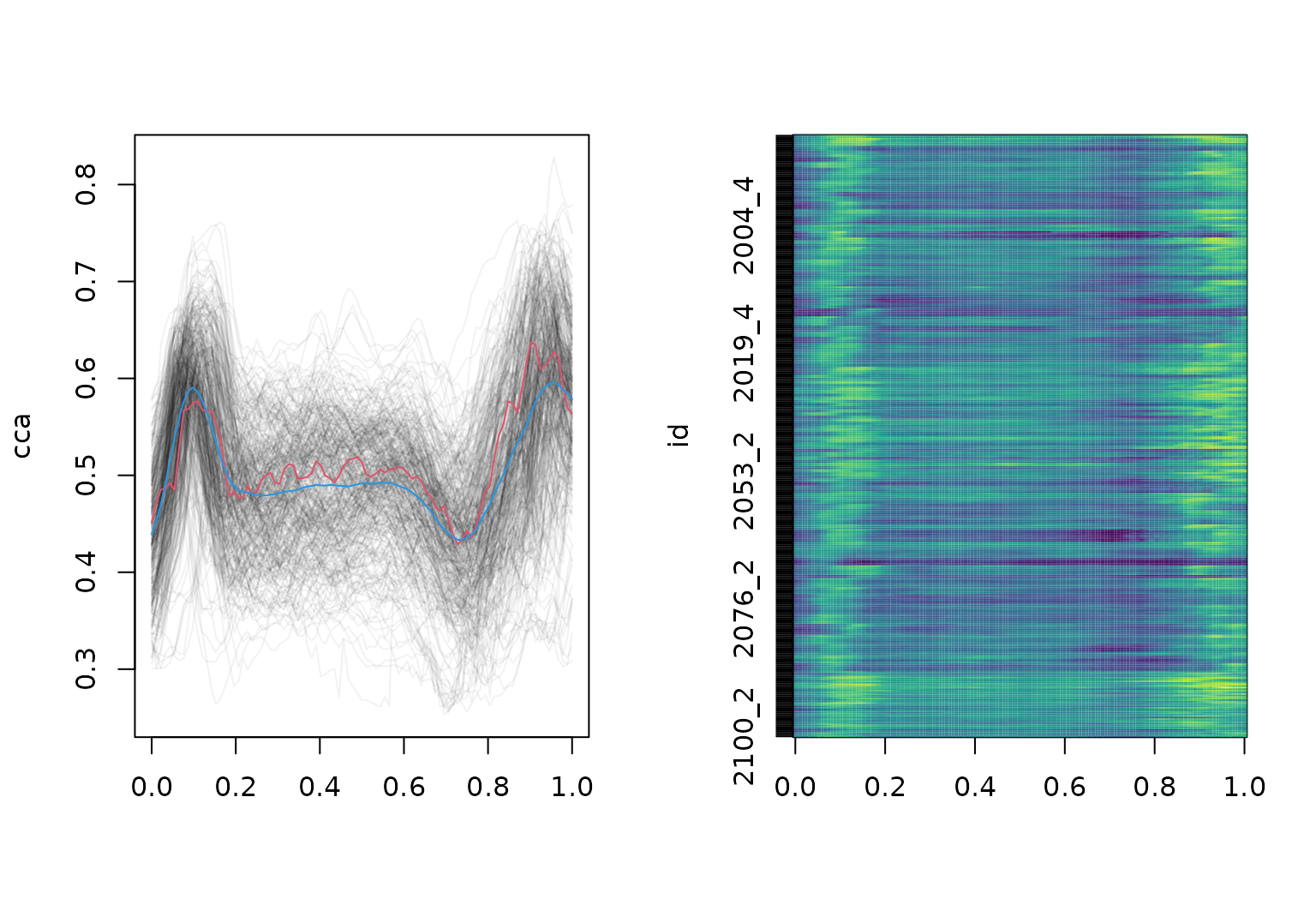
These plot methods use all the same graphics options and
can be edited like other base graphics:
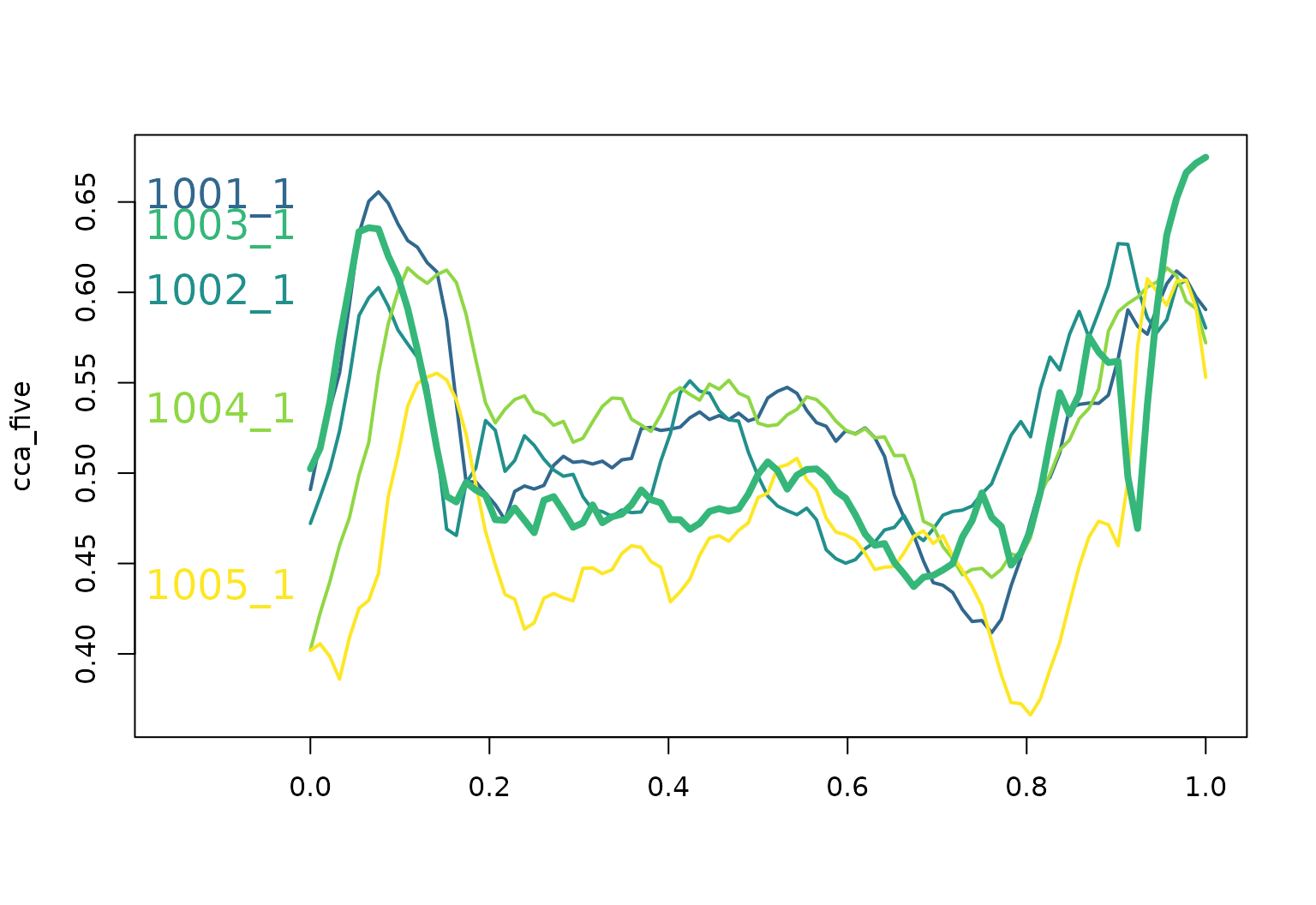
cca_five <- cca[1:5]
cca_five |> plot(xlim = c(-0.15, 1), col = pal_5, lwd = 2)
text(
x = -0.1, y = cca_five[, 0.07], labels = names(cca_five), col = pal_5, cex = 1.5
)
median(cca_five) |> lines(col = pal_5[3], lwd = 4)