Converting to & from `tf`
Jeff Goldsmith, Fabian Scheipl
2024-02-23
Source:vignettes/x02_Conversion.Rmd
x02_Conversion.RmdFunctional data have often been stored in matrices or data frames. Although these structures have sufficed for some purposes, they are cumbersome or impossible to use with modern tools for data wrangling.
In this vignette, we illustrate how to convert data from common
structures to tf objects. Throughout, functional data
vectors are stored as columns in a data frame to facilitate subsequent
wrangling and analysis.
Conversion from matrices
One of the most common structures for storing functional data has been a matrix. Especially when subjects are observed over the same (regular or irregular) grid, it is natural to observations on a subject in rows (or columns) of a matrix. Matrices, however, are difficult to wrangle along with data in a data frame, leading to confusing and easy-to-break subsetting across several objects.
In the following examples, we’ll use tfd to get a
tf vector from matrices. The tfd function
expects data to be organized so that each row is the functional
observation for a single subject. It’s possible to focus only on the
resulting tf vector, but in keeping with the broader goals
of tidyfun we’ll add these as columns to a data frame.
The DTI data in the refund package has been
a popular example in functional data analysis. In the code below, we
create a data frame (or tibble) containing scalar
covariates, and then add columns for the cca and
rcst track profiles. This code was used to create the
tidyfun::dti_df dataset included in the package.
dti_df <- tibble(
id = refund::DTI$ID,
visit = refund::DTI$visit,
sex = refund::DTI$sex,
case = factor(ifelse(refund::DTI$case, "MS", "control"))
)
dti_df$cca <- tfd(refund::DTI$cca, arg = seq(0, 1, length.out = 93))
dti_df$rcst <- tfd(refund::DTI$rcst, arg = seq(0, 1, length.out = 55))In tfd, the first argument is a matrix; arg
defines the grid over which functions are observed. The output of
tfd is a vector, which we include in the
dti_df data frame.
dti_df
## # A tibble: 382 × 6
## id visit sex case cca
## <dbl> <int> <fct> <fct> <tfd_irrg>
## 1 1001 1 female control [1]: (0.000,0.49);(0.011,0.52);(0.022,0.54); ...
## 2 1002 1 female control [2]: (0.000,0.47);(0.011,0.49);(0.022,0.50); ...
## 3 1003 1 male control [3]: (0.000,0.50);(0.011,0.51);(0.022,0.54); ...
## 4 1004 1 male control [4]: (0.000,0.40);(0.011,0.42);(0.022,0.44); ...
## 5 1005 1 male control [5]: (0.000,0.40);(0.011,0.41);(0.022,0.40); ...
## 6 1006 1 male control [6]: (0.000,0.45);(0.011,0.45);(0.022,0.46); ...
## 7 1007 1 male control [7]: (0.000,0.55);(0.011,0.56);(0.022,0.56); ...
## 8 1008 1 male control [8]: (0.000,0.45);(0.011,0.48);(0.022,0.50); ...
## 9 1009 1 male control [9]: (0.000,0.50);(0.011,0.51);(0.022,0.52); ...
## 10 1010 1 male control [10]: (0.000,0.46);(0.011,0.47);(0.022,0.48); ...
## # ℹ 372 more rows
## # ℹ 1 more variable: rcst <tfd_irrg>Finally, we’ll make a quick spaghetti plot to illustrate that the
complete functional data is included in each tf column.
dti_df |>
ggplot() +
geom_spaghetti(aes(y = cca, col = case, alpha = 0.2 + 0.4 * (case == "control"))) +
facet_wrap(~sex) +
scale_alpha(guide = "none", range = c(0.2, 0.4))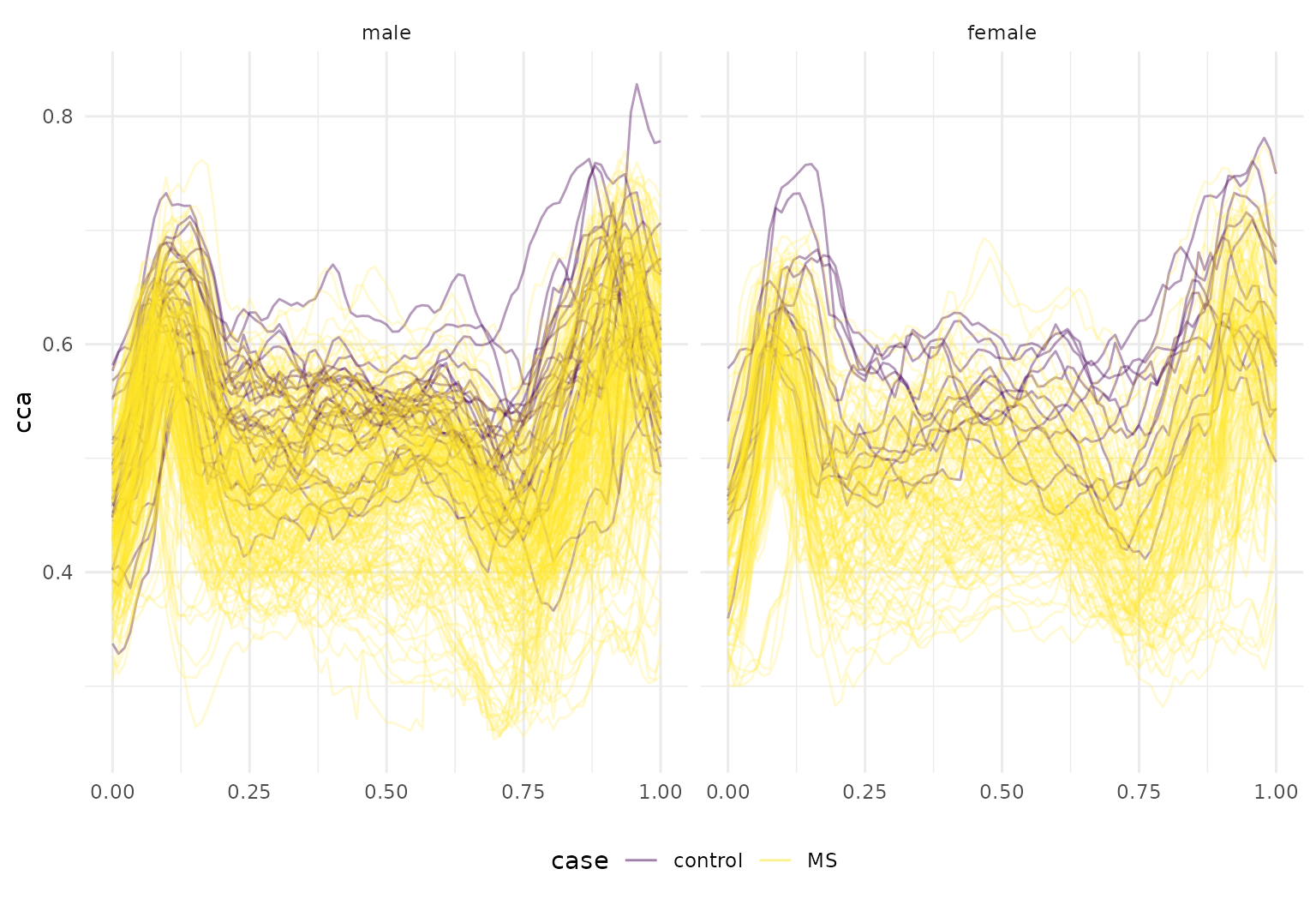
We’ll repeat the same basic process using a second, and probably
even-more-perennial, functional data example: the Canadian weather data
in the fda package. Here, functional data are stored in a
three-dimensional array, with dimensions corresponding to day, station,
and outcome (temperature, precipitation, and log10 precipitation).
In the following, we first create a tibble with scalar
covariates, then use tfd to create functional data vectors,
and finally include the resulting vectors in the dataframe. In this
case, our args are days of the year, and we use
tf_smooth to smooth the precipitation outcome. Because the
original data matrices record the different observations in the columns
instead of the rows, we have to use their transpose in the call to
tfd:
canada <- tibble(
place = fda::CanadianWeather$place,
region = fda::CanadianWeather$region,
lat = fda::CanadianWeather$coordinates[, 1],
lon = -fda::CanadianWeather$coordinates[, 2]
) |>
mutate(
temp = t(fda::CanadianWeather$dailyAv[, , 1]) |>
tfd(arg = 1:365),
precipl10 = t(fda::CanadianWeather$dailyAv[, , 3]) |>
tfd(arg = 1:365) |>
tf_smooth()
)
## using f = 0.15 as smoother span for lowessThe resulting data frame is shown below.
canada
## # A tibble: 35 × 6
## place region lat lon temp
## <chr> <chr> <dbl> <dbl> <tfd_reg>
## 1 St. Johns Atlantic 47.3 -52.4 [1]: (1, -4);(2, -3);(3, -3); ...
## 2 Halifax Atlantic 44.4 -63.4 [2]: (1, -4);(2, -4);(3, -5); ...
## 3 Sydney Atlantic 46.1 -60.1 [3]: (1, -4);(2, -4);(3, -5); ...
## 4 Yarmouth Atlantic 43.5 -66.1 [4]: (1, -1);(2, -2);(3, -2); ...
## 5 Charlottvl Atlantic 42.5 -80.2 [5]: (1, -6);(2, -6);(3, -7); ...
## 6 Fredericton Atlantic 45.6 -66.4 [6]: (1, -8);(2, -8);(3, -9); ...
## 7 Scheffervll Atlantic 54.5 -64.5 [7]: (1,-22);(2,-23);(3,-23); ...
## 8 Arvida Atlantic 48.3 -71.1 [8]: (1,-14);(2,-14);(3,-15); ...
## 9 Bagottville Atlantic 48.2 -70.5 [9]: (1,-15);(2,-15);(3,-16); ...
## 10 Quebec Atlantic 46.5 -71.1 [10]: (1,-11);(2,-11);(3,-12); ...
## # ℹ 25 more rows
## # ℹ 1 more variable: precipl10 <tfd_reg>A plot containing both functional observations is shown below.
temp_panel <- canada |>
ggplot(aes(y = temp, color = region)) +
geom_spaghetti()
precip_panel <- canada |>
ggplot(aes(y = precipl10, color = region)) +
geom_spaghetti()
gridExtra::grid.arrange(temp_panel, precip_panel, nrow = 1)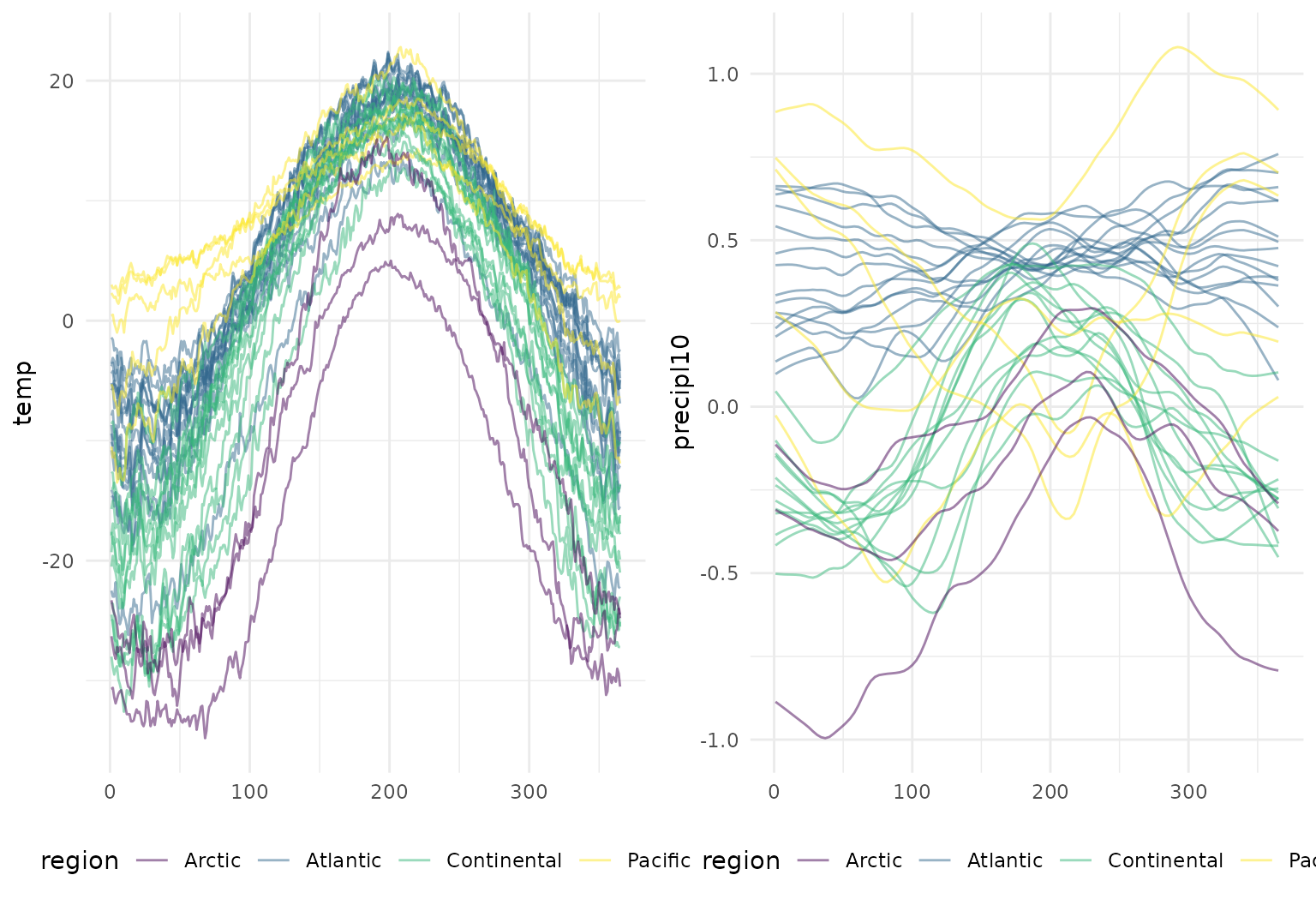
Conversion to tf from a data frame
… in “long” format
“Long” format data frames containing functional data include columns
containing a subject identifier, the functional argument, and the value
each subject’s function takes at each argument. There are also often
(but not always) non-functional covariates that are repeated within a
subject. For data in this form, we use tf_nest to produce a
data frame containing a single row for each subject.
A first example is the pig weight data from the SemiPar
package, which is a nice example from longitudinal data analysis. This
includes columns for id.num, num.weeks, and
weight – which correspond to the subject, argument, and
value.
data("pig.weights", package = "SemiPar")
pig.weights <- as_tibble(pig.weights)
pig.weights
## # A tibble: 432 × 3
## id.num num.weeks weight
## <int> <int> <dbl>
## 1 1 1 24
## 2 1 2 32
## 3 1 3 39
## 4 1 4 42.5
## 5 1 5 48
## 6 1 6 54.5
## 7 1 7 61
## 8 1 8 65
## 9 1 9 72
## 10 2 1 22.5
## # ℹ 422 more rowsWe create pig_df by nesting weight within subject. The
result is a data frame containing a single row for each pig, and columns
for id.num and the weight function.
pig_df <- pig.weights |>
tf_nest(weight, .id = id.num, .arg = num.weeks)
pig_df
## # A tibble: 48 × 2
## id.num weight
## <int> <tfd_reg>
## 1 1 [1]: (1,24);(2,32);(3,39); ...
## 2 2 [2]: (1,22);(2,30);(3,40); ...
## 3 3 [3]: (1,22);(2,28);(3,36); ...
## 4 4 [4]: (1,24);(2,32);(3,40); ...
## 5 5 [5]: (1,24);(2,32);(3,37); ...
## 6 6 [6]: (1,23);(2,30);(3,36); ...
## 7 7 [7]: (1,22);(2,28);(3,36); ...
## 8 8 [8]: (1,24);(2,30);(3,38); ...
## 9 9 [9]: (1,20);(2,28);(3,33); ...
## 10 10 [10]: (1,26);(2,32);(3,40); ...
## # ℹ 38 more rowsWe’ll make a quick plot to show the result.
pig_df |>
ggplot(aes(y = weight)) +
geom_spaghetti()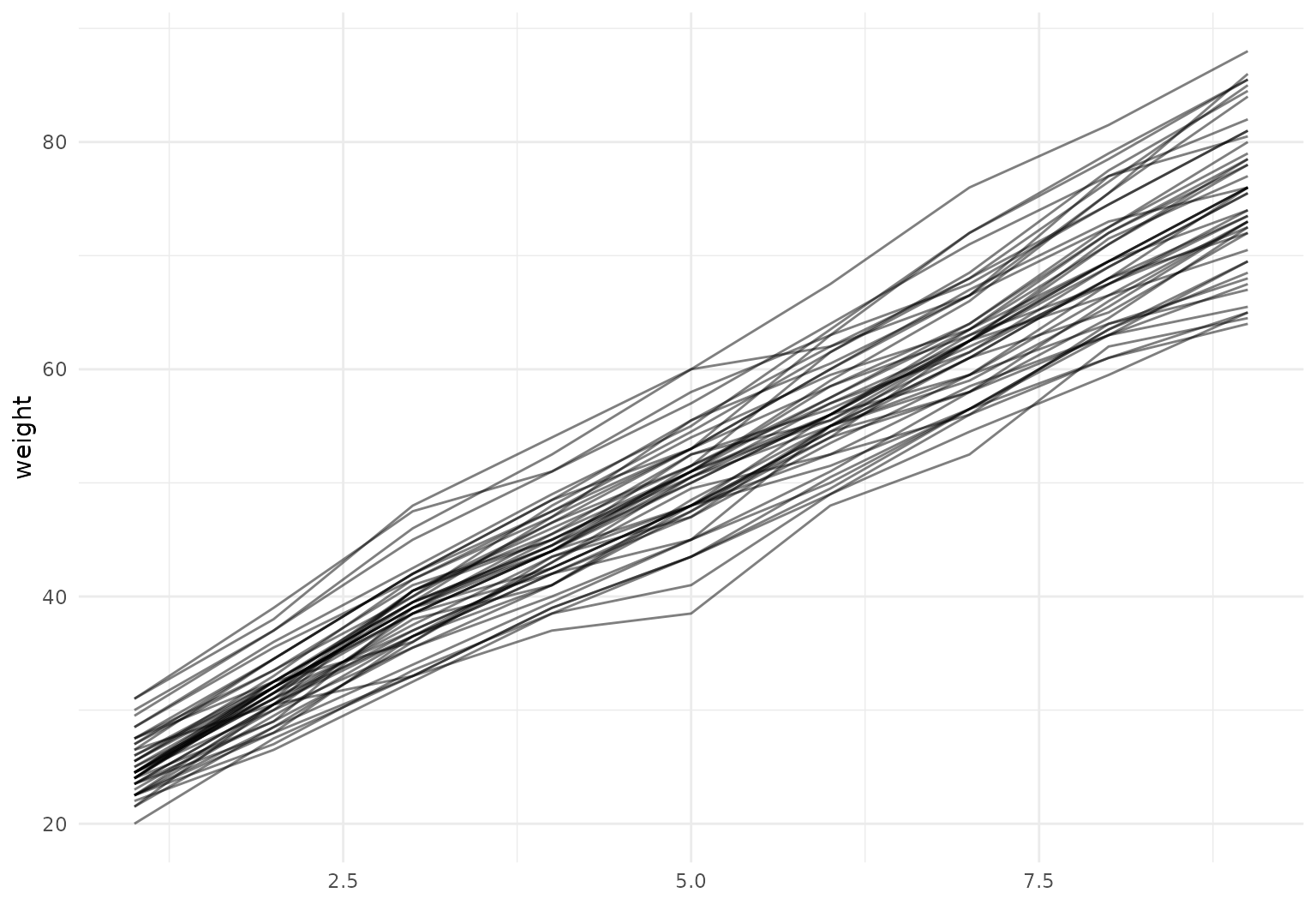
A second example uses the ALA::fev1 dataset.
ALA is not available on CRAN but can be installed using the
line below.
install.packages("ALA", repos = "http://R-Forge.R-project.org")In this dataset, both height and logFEV1
are observed at multiple ages for each child; that is, there are two
functions observed simultaneously, over a shared argument. We can use
tf_nest to create a dataframe with a single row for each
subject, which includes both non-functional covariates (like age and
height at baseline), and functional observations logFEV1
and height.
ALA::fev1 |> glimpse()
## Rows: 1,994
## Columns: 6
## $ id <fct> 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2, 3, 3, 3, 3, 3, 3, 3, 3, 3, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 5, 5, …
## $ age <dbl> 9.3415, 10.3929, 11.4524, 12.4600, 13.4182, 15.4743, 16.3723, 6.5873, 7.6496, 12.7392, 13.7741, 14.6940, 15.…
## $ height <dbl> 1.20, 1.28, 1.33, 1.42, 1.48, 1.50, 1.52, 1.13, 1.19, 1.49, 1.53, 1.55, 1.56, 1.57, 1.57, 1.18, 1.23, 1.30, …
## $ age0 <dbl> 9.3415, 9.3415, 9.3415, 9.3415, 9.3415, 9.3415, 9.3415, 6.5873, 6.5873, 6.5873, 6.5873, 6.5873, 6.5873, 6.58…
## $ height0 <dbl> 1.20, 1.20, 1.20, 1.20, 1.20, 1.20, 1.20, 1.13, 1.13, 1.13, 1.13, 1.13, 1.13, 1.13, 1.13, 1.18, 1.18, 1.18, …
## $ logFEV1 <dbl> 0.21511, 0.37156, 0.48858, 0.75142, 0.83291, 0.89200, 0.87129, 0.30748, 0.35066, 0.75612, 0.86710, 1.04732, …
ALA::fev1 |>
group_by(id) |>
mutate(n_obs = n()) |>
filter(n_obs > 1) |>
tf_nest(logFEV1, height, .arg = age) |>
glimpse()
## Rows: 252
## Columns: 6
## $ id <fct> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 13, 15, 16, 18, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 3…
## $ age0 <dbl> 9.3415, 6.5873, 6.9131, 6.7598, 6.5024, 6.8994, 6.4339, 7.1869, 6.8966, 7.7892, 7.6140, 7.5483, 7.8412, 6.50…
## $ height0 <dbl> 1.20, 1.13, 1.18, 1.15, 1.11, 1.24, 1.18, 1.27, 1.17, 1.13, 1.32, 1.25, 1.25, 1.20, 1.19, 1.24, 1.21, 1.23, …
## $ n_obs <int> 7, 8, 9, 10, 7, 11, 7, 9, 9, 10, 6, 3, 5, 11, 12, 10, 9, 8, 12, 2, 2, 11, 11, 7, 9, 11, 11, 4, 2, 12, 3, 9, …
## $ logFEV1 <tfd_irrg> [<9.3415, 10.3929, 11.4524, 12.4600, 13.4182, 15.4743, 16.3723>, <0.21511, 0.37156, 0.48858, 0.75142, 0…
## $ height <tfd_irrg> [<9.3415, 10.3929, 11.4524, 12.4600, 13.4182, 15.4743, 16.3723>, <1.20, 1.28, 1.33, 1.42, 1.48, 1.50, 1…… in “wide” format
In some cases functional data are stored in “wide” format, meaning
that there are separate columns for each argument, and values are stored
in these columns. In this case, tf_gather can be use to
collapse across columns to produce a function for each subject.
The example below again uses the refund::DTI dataset. We
use tf_gather to transfer the cca observations
from a matrix column (with NAs) into a column of
irregularly observed functions (tfd_irreg).
dti_df <- refund::DTI |>
janitor::clean_names() |>
select(-starts_with("rcst")) |>
glimpse()
## Rows: 382
## Columns: 8
## $ id <dbl> 1001, 1002, 1003, 1004, 1005, 1006, 1007, 1008, 1009, 1010,…
## $ visit <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
## $ visit_time <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
## $ nscans <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
## $ case <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
## $ sex <fct> female, female, male, male, male, male, male, male, male, m…
## $ pasat <int> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
## $ cca <dbl[,93]> <matrix[26 x 93]>
dti_df |>
tf_gather(starts_with("cca")) |>
glimpse()
## creating new tfd-column <cca>
## Rows: 382
## Columns: 8
## $ id <dbl> 1001, 1002, 1003, 1004, 1005, 1006, 1007, 1008, 1009, 1010,…
## $ visit <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
## $ visit_time <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
## $ nscans <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
## $ case <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
## $ sex <fct> female, female, male, male, male, male, male, male, male, m…
## $ pasat <int> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
## $ cca <tfd_irrg> [<1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 1…Reversing the conversion
tidyfun includes a wide range of tools
for exploratory analysis and visualization, but many analysis approaches
require data to be stored in more traditional formats. Several functions
are available to aid in this conversion.
Conversion from tf to data frames
The functions tf_unnest and tf_spread
reverse the operations in tf_nest and
tf_gather, respectively – that is, they take a data frame
with a functional observation and produce long or wide data frames.
We’ll illustrate these with the pig_df data set.
First, to produce a long-format data frame, one can use
tf_unnest:
pig_df |>
tf_unnest(cols = weight) |>
glimpse()
## Rows: 432
## Columns: 3
## $ id.num <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2, 2, 3, …
## $ weight_arg <int> 1, 2, 3, 4, 5, 6, 7, 8, 9, 1, 2, 3, 4, 5, 6, 7, 8, 9, 1, …
## $ weight_value <dbl> 24.0, 32.0, 39.0, 42.5, 48.0, 54.5, 61.0, 65.0, 72.0, 22.…To produce a wide-format data frame, one can use
tf_spread:
pig_df |>
tf_spread() |>
glimpse()
## Rows: 48
## Columns: 10
## $ id.num <int> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18…
## $ weight_1 <dbl> 24.0, 22.5, 22.5, 24.0, 24.5, 23.0, 22.5, 23.5, 20.0, 25.5, 2…
## $ weight_2 <dbl> 32.0, 30.5, 28.0, 31.5, 31.5, 30.0, 28.5, 30.5, 27.5, 32.5, 3…
## $ weight_3 <dbl> 39.0, 40.5, 36.5, 39.5, 37.0, 35.5, 36.0, 38.0, 33.0, 39.5, 4…
## $ weight_4 <dbl> 42.5, 45.0, 41.0, 44.5, 42.5, 41.0, 43.5, 41.0, 39.0, 47.0, 4…
## $ weight_5 <dbl> 48.0, 51.0, 47.5, 51.0, 48.0, 48.0, 47.0, 48.5, 43.5, 53.0, 5…
## $ weight_6 <dbl> 54.5, 58.5, 55.0, 56.0, 54.0, 51.5, 53.5, 55.0, 49.0, 58.5, 5…
## $ weight_7 <dbl> 61.0, 64.0, 61.0, 59.5, 58.0, 56.5, 59.5, 59.5, 54.5, 63.0, 6…
## $ weight_8 <dbl> 65.0, 72.0, 68.0, 64.0, 63.0, 63.5, 67.5, 66.5, 59.5, 69.5, 6…
## $ weight_9 <dbl> 72.0, 78.0, 76.0, 67.0, 65.5, 69.5, 73.5, 73.0, 65.0, 76.0, 7…Converting back to a matrix or data frame
To convert tf vector to a matrix with each row
containing the function evaluations for one function, use
as.matrix:
weight_vec <- pig_df$weight
weight_matrix <- weight_vec |> as.matrix()
head(weight_matrix)
## 1 2 3 4 5 6 7 8 9
## 1 24.0 32.0 39.0 42.5 48.0 54.5 61.0 65.0 72.0
## 2 22.5 30.5 40.5 45.0 51.0 58.5 64.0 72.0 78.0
## 3 22.5 28.0 36.5 41.0 47.5 55.0 61.0 68.0 76.0
## 4 24.0 31.5 39.5 44.5 51.0 56.0 59.5 64.0 67.0
## 5 24.5 31.5 37.0 42.5 48.0 54.0 58.0 63.0 65.5
## 6 23.0 30.0 35.5 41.0 48.0 51.5 56.5 63.5 69.5
# argument values of input data saved in `arg`-attribute:
attr(weight_matrix, "arg")
## [1] 1 2 3 4 5 6 7 8 9To convert a tf vector to a standalone data frame with
"id","arg","value"-columns, use
as.data.frame() with unnest = TRUE:
weight_vec
## tfd[48] on (1,9) based on 9 evaluations each
## interpolation by tf_approx_linear
## 1: (1,24);(2,32);(3,39); ...
## 2: (1,22);(2,30);(3,40); ...
## 3: (1,22);(2,28);(3,36); ...
## 4: (1,24);(2,32);(3,40); ...
## 5: (1,24);(2,32);(3,37); ...
## [....] (43 not shown)
weight_vec |>
as.data.frame(unnest = TRUE) |>
head()
## id arg value
## 1.1 1 1 24.0
## 1.2 1 2 32.0
## 1.3 1 3 39.0
## 1.4 1 4 42.5
## 1.5 1 5 48.0
## 1.6 1 6 54.5